super-resolution ribosome profiling
Ribosome profiling (Ribo-seq) enables visualization of ribosome positions
on individual mRNA molecules. The general procedure involves nuclease
digestion, size selection, cDNA construction, and deep sequencing.
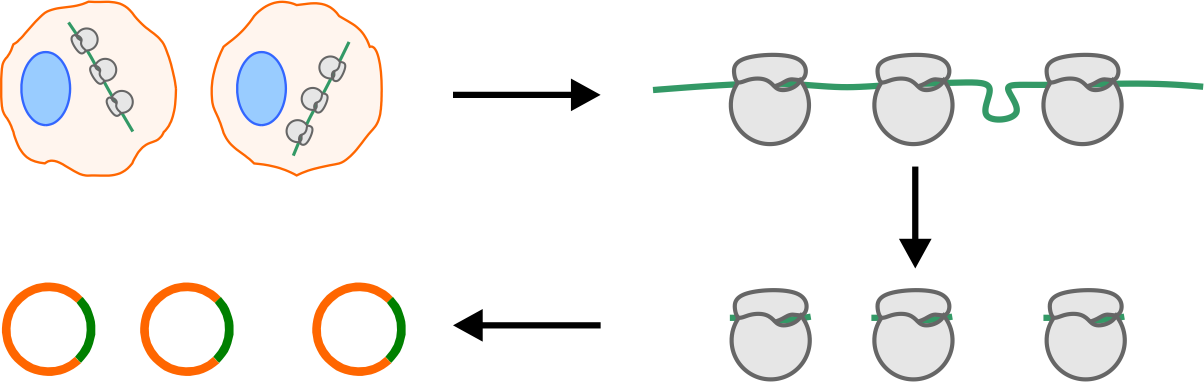
Current ribosome profiling involves circularization-based methodology
for cDNA synthesis, which has low efficiency. Additionally, the ligation
introduces untemplated nucleotides. Typical RNA ligation suffers from
sequence-dependent biases, resulting in altered footprint quantification.
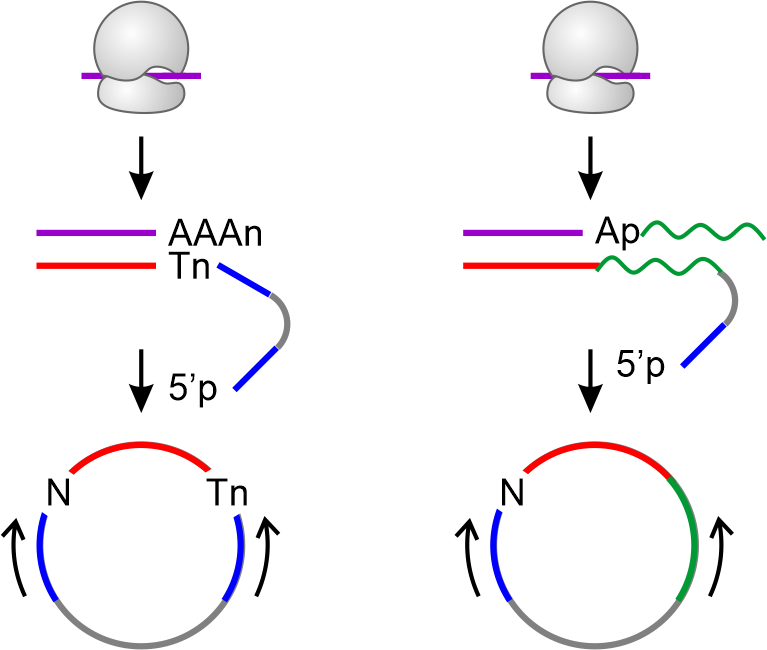
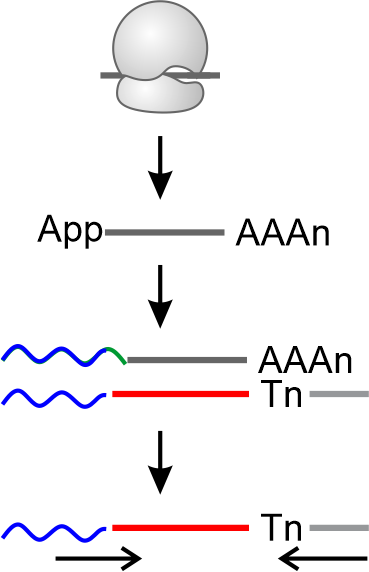
QEZ-seq ® introduces proprietary RNA adenylation technology, enabling
5' end adenynylation and 3' end polyadenylation. Without 3' end linker ligation
and 5' end circularization, Ezra-seq simplifies the library construction.
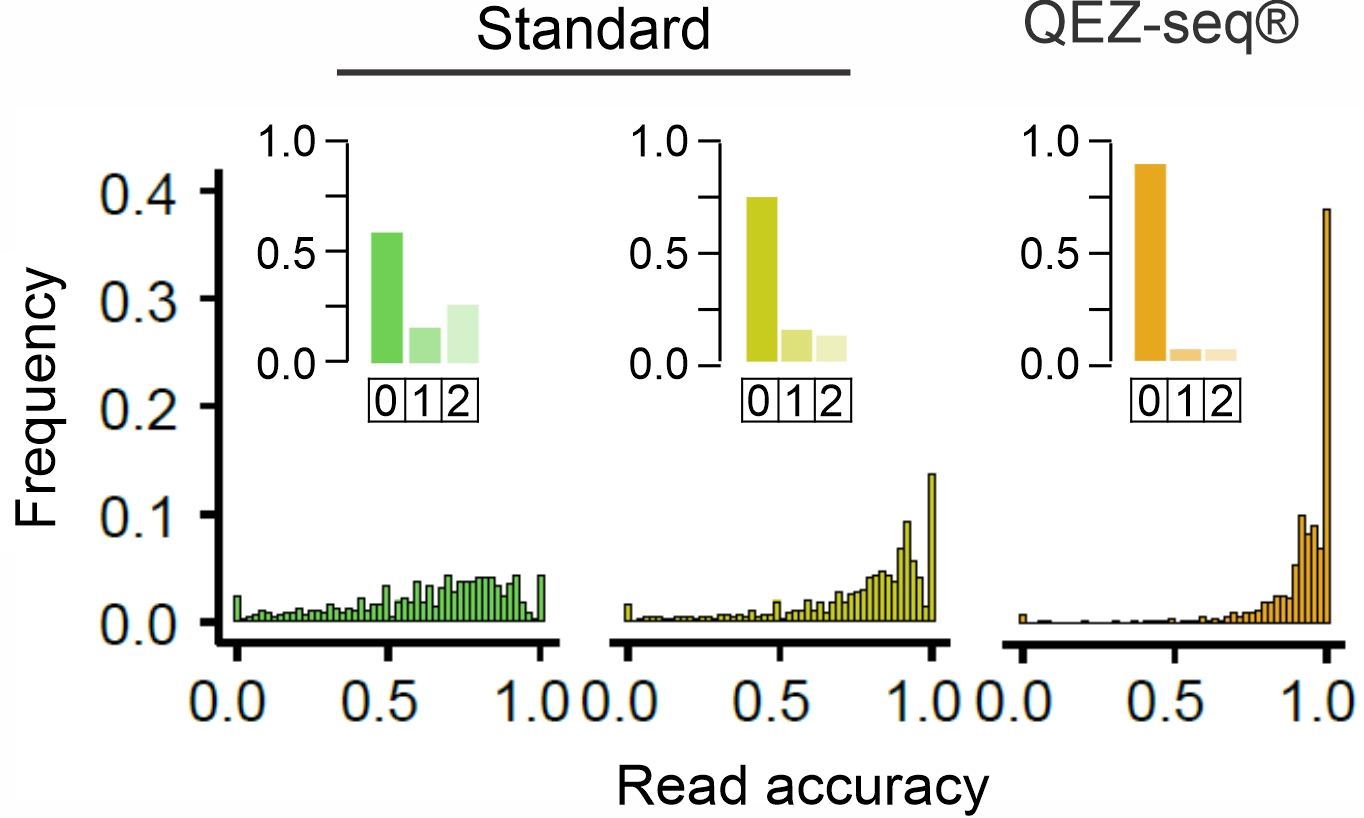
QEZ-seq ® improvies the mapping accuracy of ribosome footprints.
The ribosome positions inferred form the 5' end of footprints are independent of the read length.